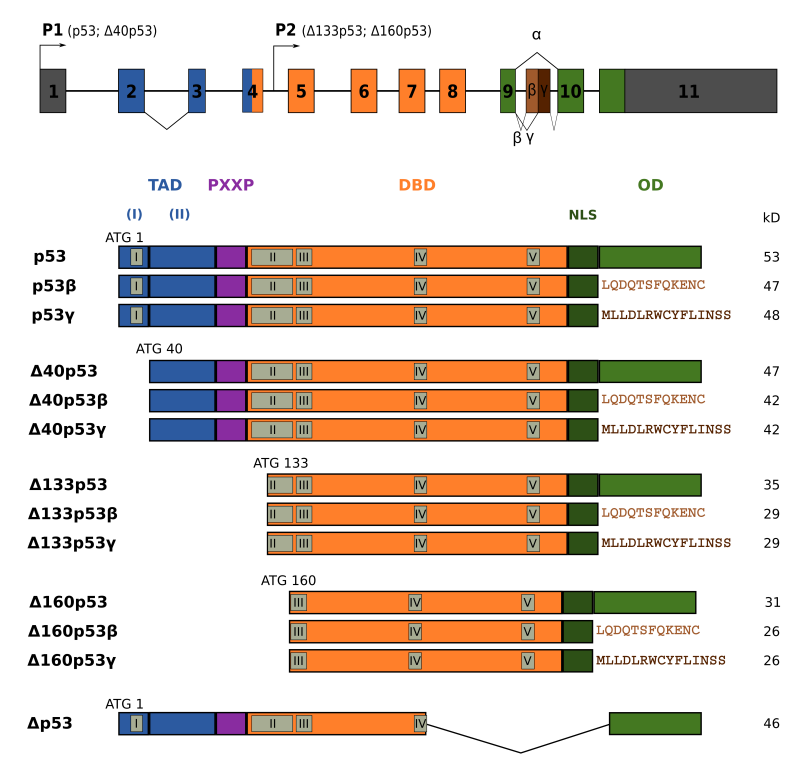
Several isoforms of p53 have been described that are produced by alternative splicing, use of alternative translation site or alternative promoter (see figure below and Marcel & Hainaut, 2008, Marcel et al., 2011).
In the IARC TP53 Database, all annotations and data are in relation to the full-lenght protein (p53 or p53TA isoform). Annotations were generated on the predicted impact of TP53 gene mutations on the protein status of isoforms.
All mutations recorded in the database have thus been analyzed to predict their consequences on the
status of each p53 protein isoform.
These predictions are based on theoretical approaches taking into account the known protein sequences of
each isoforms, the position of
the mutation in these sequences and the type of variation.
Annotations and rules used for the predictions:
- WT = no amino acid change compared to the wild-type sequence
- Altered = any change in the amino acid sequence
- Not produced = any mutation affecting the start codon of the isoform
- Unknown = uncertain prediction of variation effect
These annotations can be retrieved from the 'Functional / Structural Data' search option.
- p53alpha: NM_000546
- p53beta: DQ186648, Bourdon et al., 2005.
- p53gamma: DQ186649, Bourdon et al., 2005.
- delta40p53alpha: Courtois et al, 2002.
- delta40p53beta: Bourdon et al., 2005.
- deltap4053gamma: Bourdon et al., 2005.
- delta133p53alpha: DQ186650, Bourdon et al., 2005.
- delta133p53beta: DQ186651, Bourdon et al., 2005.
- delta133p53gamma: DQ186652, Bourdon et al., 2005.
- deltap53: Rohaly et al., 2005